Transposable DNA element | Classification of Transposable Elements (TEs)
Introduction: The transposable elements are those vital genetic elements that can move from one position to another within a genome or chromosome or plasmid and creating a genetic instability. It was first discovered by American plant geneticist Barbara Mcclintock and published in her famous publication in 1948, when deoxyribonuclic acid (DNA) was not even established as a genetic material. She got Nobel Prize in 1983 for her great work. Occasionally transposable elements.
(TE) are responsible for altering cell`s genome size by creating or reversing mutations. The transposing of a gene usually occurs by the duplication of the respective transposable elements (TE) . Some scientists mentioned them as jumping genes.
Transposable elements (TE) have been observed in all types of organisms both prokaryotic and eukaryotic cells. For example, TEs can be found approximately:
| in several fish species | 10% |
| in genom of C. Elegans | 12% |
| In genom of mouse | 37% |
| In human genome | 45% |
| In maize genome | upto > 80% |
Even small organism bacteria to large organism human shows transposable elements that have been compiled over time and being continued to make a new shape of genomes through the process of their mobilization.
The movement of TEs can be treated as transposition or retrotransposition which is depend on the types of intermediate which is used in mobilization process .There are various procedures of activities of TEs which shows positive or negative impact on genome size . As per example, TEs mobilization can be increase the gene activation, control gene expression, induce improper recombination. In such a way, TEs play vital role in genome evolution. Although some theoretical perspectives TEs are termed as selfish DNA or junk DNA. So when these elements are present in a genome make conflicting among the selfish DNA ( in a conditon of perpetuated) and respective host ( to shorten it’s spread and consequences).
As Transposable elements are made up big percentage of genome volume, so it can be assumed that these elements can play a vital role in changing genome size by the process of speciation and evolution and it is shown in both plants and animals or insects like Drosophila and primates also . As the trigger(s) using in transposable elements-induced genome size promotes is still clearly unknown, so it is hypothesized that stress condition can be implicated during the amplification of transposable elements. Transposable elements are involved for producing different types of genetic alterations to make successful transposition process for example, excisions, translocation , insertions, duplications in the site of integration . Even DNA transponson could be inactivate or alter the gene expression by insertion process within the introns, exons or in regulatory regions.
[13].TEs are also able for making up a big fraction of genome and responsible for C- value of eukaryotic cells. So TEs are very significant for genomic function and evolution.
[14.].Oxytricha is an exclusive genetic system where TEs play an important role for development [15]. However two classes of TEs are available:
Class-1(retrotransposons) which is worked by the procedure of reverse transcription.
Class-2 (DNA transposons) which worked via insertion and excision and encoded by transposase.
Discovery of Transposable Elements: Nobel laureate Amrican plant geneticist Barbara Mcclintock first discovered transposable elements in Zea mays, scientific name of maize at cold spring harbor laboratory in New York, America [17]. In the year of 1944-45 (winter season) scientist Barbara Mcclintock started her experiment by planting corn kernel via self pollination . These experimental kernels were originated from long line plants and the edge of ninth chromosome was showing broken arms. She also observed some unexpected color patterns on the leaves during growing stage such as one leaf showed twice-identical albine patches on the side by side directions of that specific leaf [17]. Then scientist Mcclintock hypothesized some specific cells had lost their genetic materials, mean while remaining cells were received that they lost prior [18]. That means specific portions of a specific chromosome had switched the position. Then she also postulated that these genes were not only moved from one position to another but also turned on or off just because of specific environmental factors during various steps of cell divisions [18].

Fig 01: Bacterial composite transponse.
At the mean time she also stated that gene mutation would be reversed in this conditions.[19]. Finally, in 1951 Mcclintock presented her overall findings and in 1953 her article named “Introduction of Instability at selected lociin Maize” was published [20]. However, unfortunately her report had completely rejected and neglected at that time. But when transposable elemennts(TEs) were discovered at bacteria in 1960s-1970s then her findings were accepted by all. As a result, in 1983 she got Nobel Prize for her work in the field of physiology or medicine.
Classification of Transposable Elements (TEs): There are different types of TEs are founded in the nature like maize. But on the basis of transposition mechanism transposable element are basically two types:
- Class -1: Retrotrasposons -mainly observed in viruses like HIV virus.
- Class-2: Transposons – mainly observed in eukaryotes and prokaryotes.
Class -1(Retrotrasposons): It means transposable elements will be copied in twice steps. First of all, elements are transcribed from DNA to RNA and after that RNA again reverse transcribed to DNA. This cDNA again insertd to previous genom but in new position by the catalyzation of reverse transcriptase enzyme that is frequently encoded by transposable element itself. Class -1 are again subdivided into three groups:
- Long terminal repeats : encoded by reverse transcriptase
- Long interspersed nuclear elements: encoded by reverse transcriptase beyond long terminal repeats and transcription occurred by RNA polymerase II.
- Short interspersed nuclear elements: without encoded by reverse transcriptase but transcription occurred by RNA polymerase III.
Class -2 (Transposons): It just follows cut and paste mechanism where no need RNA as intermediate but canalization performed by several transposes enzymes. Among the different transposase enzymes particular enzymes nonspecifically bind into DNA target site whereas others are binded as specific target sequences. Briefly, here DNA is copied by transpose mechanism by the help of DNA polymerase and Dna ligase enzymes. Here also observes gene duplication and genomic evolution during S phase (Synthesis phase where DNA is replication in between G1 phase and G2 phase) of cell cycle. It occurs both eukaryotes and prokaryotes.
On the other hand, transposition elements are further classified according to their movement. These are: a) autonomous and b) non-autonomous which are both belongs to class- 1 and class-2 TEs. Autonomous transposable elements are able to move by themselves, for example activator elements (AC) but non-autonomous TEs need another TEs for their movement, for example dissociation elements (DS).
Families of Transposase: Five distinct transposes families have observed is described below:
- DDE transposases: It contains three different types of amino acid: aspartate (D), aspartate (D) and glutamate (E) which is needed to cordinate with metal ion for catalyzation and DDE is very much integrated in diffrent ways of transposition cycle. Typical example of this family is observed in drosophila pelement , bacteriophage Mu, Tn5, and Mainer , IS10 and Is50.
- Tyrosine(Y) transposases: This family has shown in cut and paste mechanism of transposition. It is usullly observed in bacteria, kangaroo, Tn916 and DIRS1.
- Serine transposases: This family is also involved in cut and paste mechanism of transposition especially in circular DNA intermediate of site specific serine residue. This family usually involved in bacteria, Tn5397 and IS607.
- Rolling –circle(RC) or Y2 transposases: This family is involved in copy-in mechanism where they help to copy a single strand DNA into the replicated DNA’s target site. As a result newly synthesized strand has formed by the help the template DNA and cDNA.It is observed in IS91 and helitron .
- Reverse transcriptases or endonucleases(RT/EN): Usually they are used in different Mechanism of transposition, for example in RT/EN method. It helps to nick the target site of DNA and in reverse transcription of RNA as a nick saving primer with the help of transcriptase enzyme. It is observed in LINE-1 and TP-retrotransposons.
Examples of Transposable Elements: The existance of transposable elements are first discovered by McClintock in maize (Zea mays) genome in 1948 and she told insertions, deletion, and translocation are occured by these TEs. P elements of Drosophila melanogaster are the example of TEs .
TEs are also seen in bacteria, which is responsible for antibiotic resistance. In case of bacterial genome the transposable elements are able to jump from one chromosal DNA to plasmid DNA which can back again and able to show multi-antibiotic resistant.
TEs are also gound in human genome, which is called Alu sequence. In human genome it is almost 300 bases long and approximately found inbetween 300,000 to 10,00,000 times. Alu sequence alone contains 15-17% in human genome.
Mariner like elements is also prominent example of TEs which are found many species including human. Jacobson and Hart in Drosophila first invent it [28]. Human genome contains 14,000 copies (2.6 million base pairs) of mariner elements [29]. The first mariner-element transposons also found in human parasite named Trichomonas vaginalis.
Saccharomyces cerevisiae is a yeast which has five prominent retrotranposon families named Ty1, Ty2, Ty3, Ty4 and T.
Eukaryotes also carry TEs known as Helitron, which helps in repliation, by the mechanism of rolling circle.
General mechanism of Transposition: As American geneticist McClintock first described about transposable element in 1948 and she told that there are two moveable elements are involved in total mechanism. These moveable elements are known as activator (AC) and Dissociation (DS). So DS was only performed their activity inthe presence of AC. But intersting thing is that AC can move itself. As transposable elements are able to access into various locations of particular genome, even though within the genes due often result of mutations of those particular genes. However, DS elements are resulting from the deletion mutant of AC element.
AC elements are in generally 4.5kb in long and encoded by 3.5kb mRNA and move on by the help of transposase protien. In case of AC elements, transposase genes which are important in transposition mechanism smoothly flanked on either side by itself by 10b reverse repeat sequence known as IR. However, every IR carries an AAACGG motif, which is worked as transposase binding sequence. Thus moving ability of transposable elements would be abolished due to mutation.

Fig 02: AC Transposable elements.
As the transposable elements represents recognise sequences in the genome, which can make 8bp staggered cuts in the particular chromosome during transposition. When the cut will be staggered, mean while 8bp single stranded genomic DNA on its either side of TEs would be filled by the help of host DNA polymerase enzyme . Thus, 8bp target site duplication will be created which will flank the TEs. While TEs will be excised from the the site a bit later but 8bp duplication will still exist at that site as a direct repeat for marking where shows the evidence of existence of transposable element as a footprint.
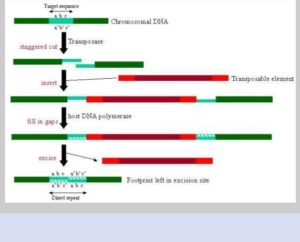 Fig 03: Transposition: transposable element inserts into staggered cuts in chromosomal DNA, the gaps being filled in by DNA polymerase. Upon excision, direct repeats are left in the chromosomal DNA.
Fig 03: Transposition: transposable element inserts into staggered cuts in chromosomal DNA, the gaps being filled in by DNA polymerase. Upon excision, direct repeats are left in the chromosomal DNA.
Diseases caused by TEs : Transposable elements are resposible diffrent types of diseases like hemophilia A and B, severe combined immunodeficiency by HIV virus, porphyria, also predisposing factor of cancer , duchenne muscular dystrophy.
Conclusion: Transposable elements are very useful tools in both plant and human molecular biology. Many researchers use the TEs as the meaning of mutagenesis. Although it makes some disease but also it is used in, beneficial purposes like to identify cancer gene for example in sleeping beauty transposing system.
Written by Md Abu Bakar Siddik, Research fellow, Department of Animal genetics and breeding.
George-August University, Göttingen, Germany.
4,336 total views, 1 views today




Like!! I blog quite often and I genuinely thank you for your information. The article has truly peaked my interest.
I like this web blog it’s a master piece! Glad I observed this
ohttps://69v.topn google.Raise your business